Summary of SCPortalen
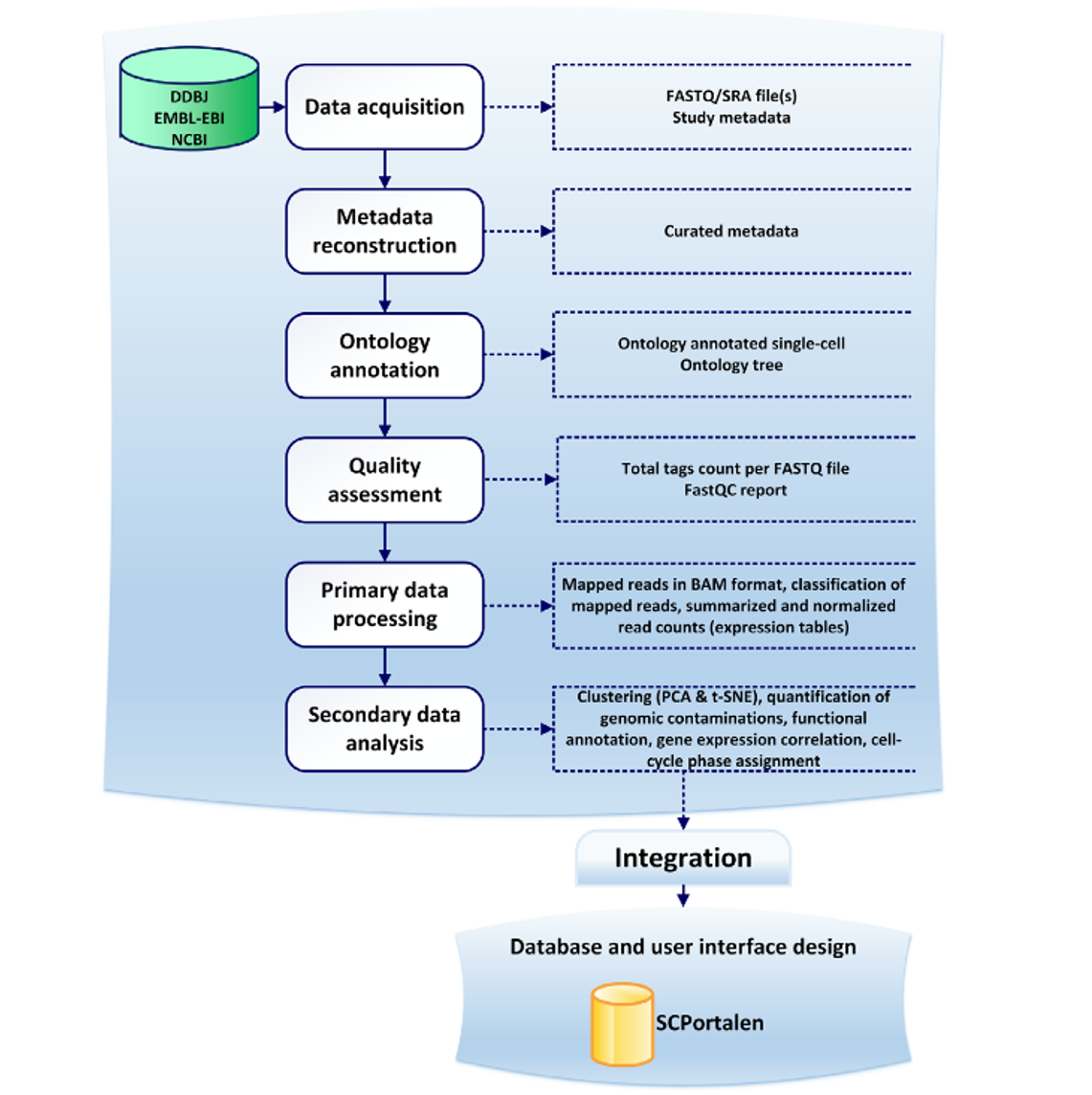
Database Model: Dataset Workflow
The following figures are taken from the SCPortalen main publication and outlines the general workflow for acquiring, processing and publishing single-cell datasets. The workflow consists of six processes. The main input to the workflow is study accession number, which allows for integration with INSDC Databases. Raw sequence files (FASTQ/SRA) and the study metadata are aquired. Followed by quality assessment procedures, metadata construction and ontology annotation. All outputs are integrated into the SCPortalen database.
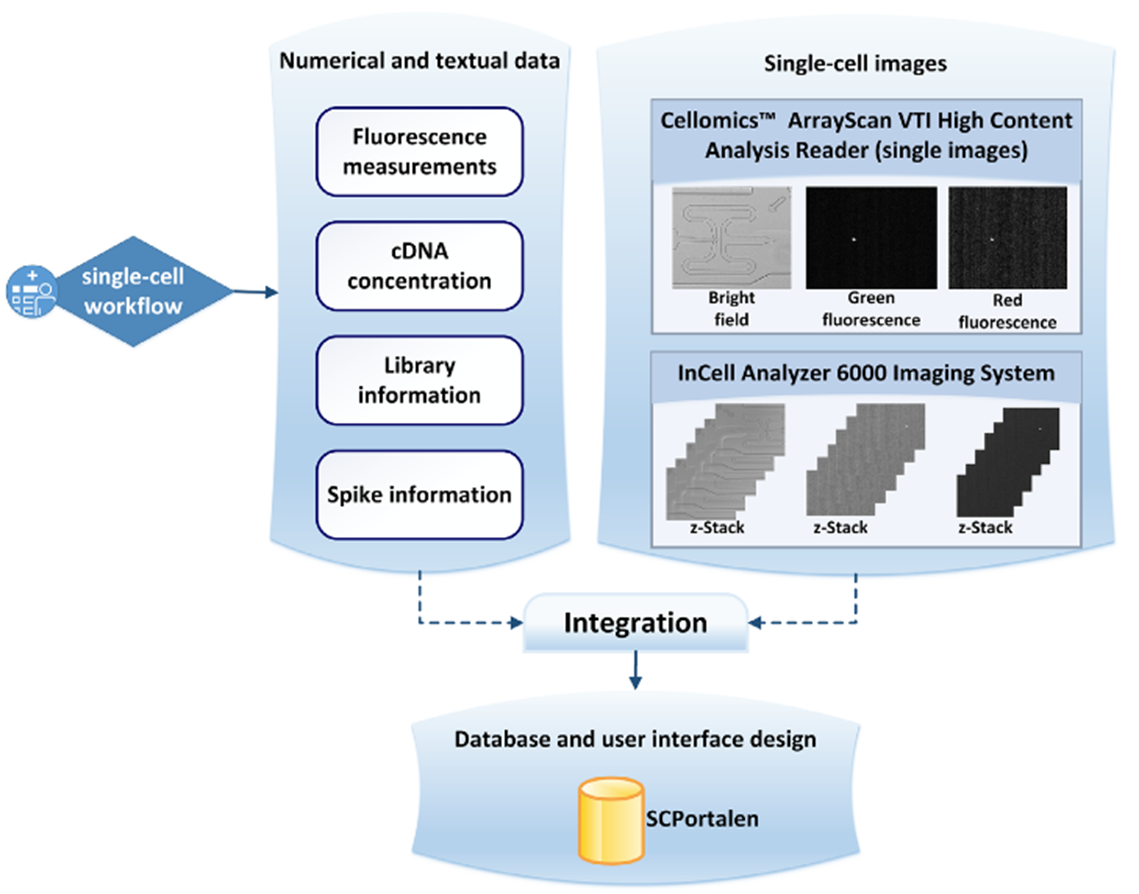
Database Model: Cell-Image Workflow
The workflow for cell-images is as detailed below. Two microscope platforms for cell-image capture are selected for integration into the database.
- CellomicsTM - Green-fluorescence, red-fluorescence and brightfield images are supplied by this platform.
- InCell Analyzer 6000 - With this platform, SCPortalen offers a movie file for each cell showing a run-through of optical sections taken. A Z-stack image of these sections is also offered.
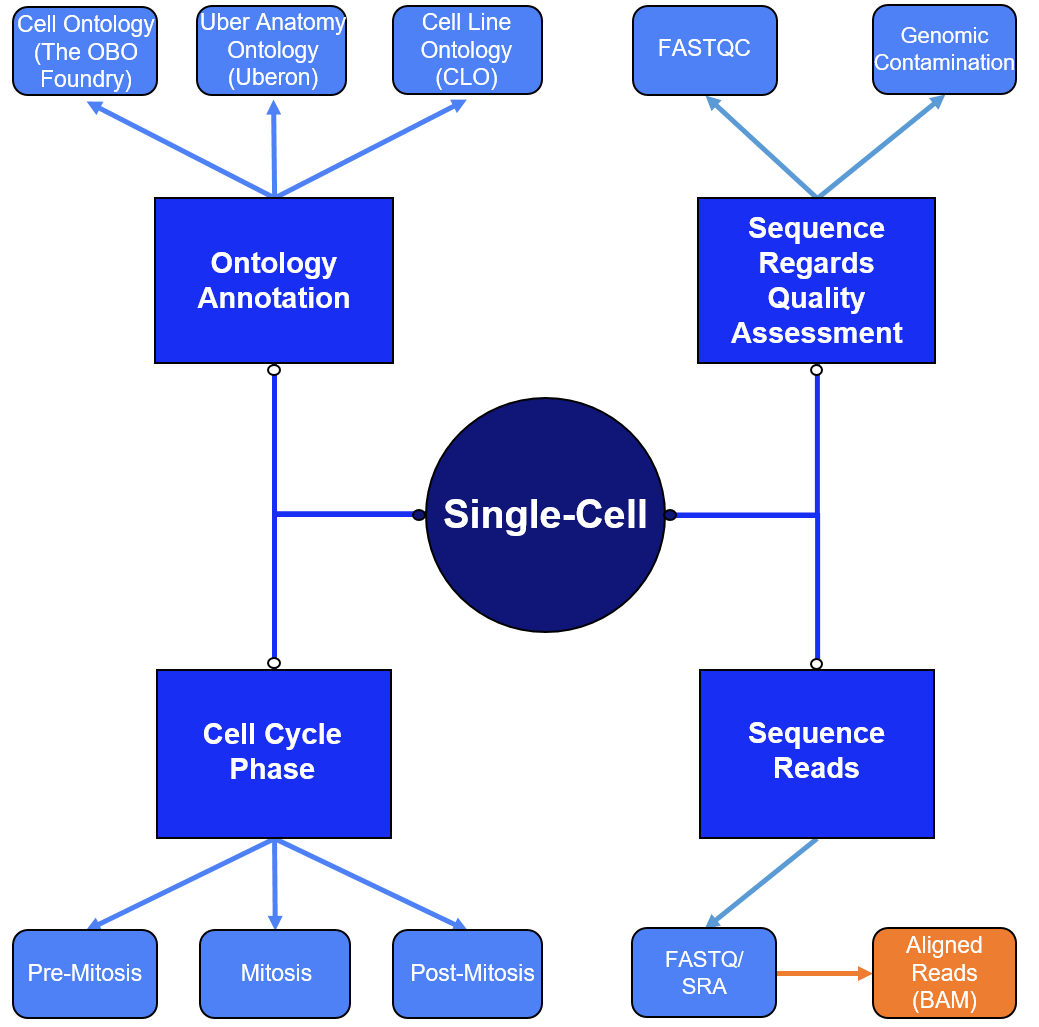
High Level
The overarching structure of data curation is outlined in the figure below. Data for cell/cell line ontology is provided in interactive flow diagrams. Strict quality control (QC) is manually curated; FASTQC reports are provided as well as particular attention to genomic contamination. Cell cycle phases are also provided.
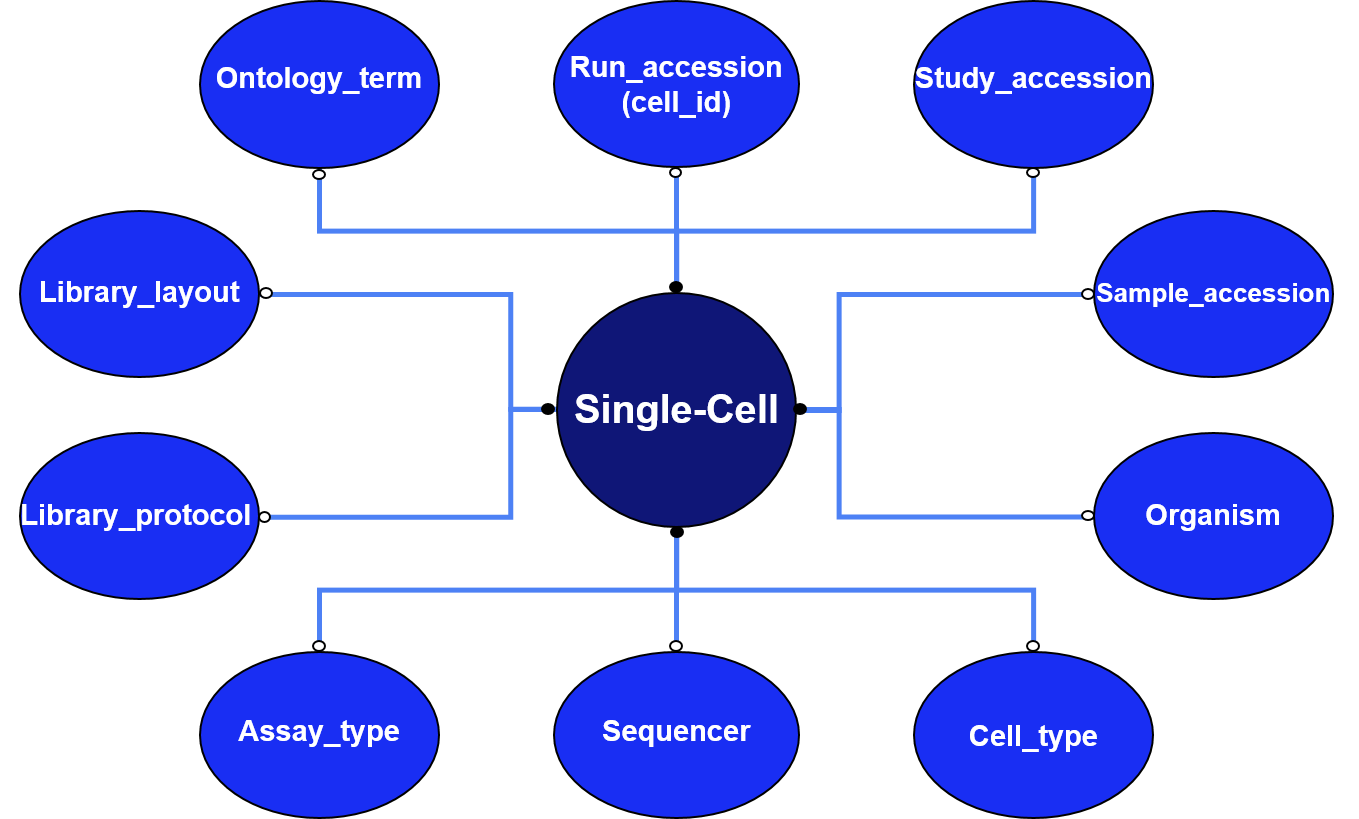
Cell Identity Metrics
Each cell is uniquely identified with integrated accession numbers at all levels (study, sample and individual run). As well as this, technical metadata information for each cell, including sequencer, assay type and library information, is given. Data from the analysis pipeline is then provided.
10X Genomics Datasets
10X Genomics' Chromium, and Cellranger analysis pipeline, is an increasingly popular protocol that retrieves huge amounts of data from 10-100s of thousands of cells to more than 1 million in some datasets. This presents a unique challenge to a cell-centric database. To accommodate and integrate data from this popular protocol in SCPortalen, we have adapted our pipeline to present this data in a cluster specific.